Abstract
Three species of sparids in the western Atlantic, sheepshead (Archosargus probatocephalus), sea bream (A. rhomboidalis), and pinfish (Lagodon rhomboides), share overlapping habitats, spawning seasons, and spawning grounds, providing opportunities for interaction among these species. Three regions of mitochondrial DNA and three nuclear DNA intron sequences were used to construct the genetic relationships among these species. The results showed that these species are closely related, suggesting the presence of soft polytomy with sheepshead and western Atlantic sea bream as sister species. However, western Atlantic sea bream and pinfish are equally divergent from sheepshead. We used a suite of 18 microsatellite markers to verify the occurrence of hybridization, identify the parental types, and evaluate the filial-generation status of 36 individuals morphologically identified as hybrids from the Indian River Lagoon system, in Florida. The 36 putative hybrids were analyzed with a reference group of 172 western Atlantic sea bream, 232 pinfish, and 157 sheepsheads and were all genetically determined to be F1 of sheepshead and western Atlantic sea bream with very little indication or no introgressive hybridization among the 172 reference specimens of western Atlantic sea bream. Hybridization was asymmetric, with western Atlantic sea bream males crossing with sheepshead females. Hybrids were first observed in the Indian River Lagoon in 2005, after the western Atlantic sea bream had become common there, in the 1990s. Their occurrence could be associated with unique features of the Indian River Lagoon that bring the two species together or with recent anthropogenic changes in this system. Further study is needed to determine the causes and long-term effects of the recurrent production of F1 hybrids and the degree of their sterility in the Indian River Lagoon.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Indian River Lagoon (IRL) system is a 156-mile-long, narrow coastal lagoon on the Atlantic coast of the Florida peninsula, USA (Fig. 1). It is relatively shallow and has relatively high salinity, with few, small, and widely scattered freshwater tributaries and few, widely scattered inlets adjoining with the nearshore marine environment (Gilmore 1995; Dybas 2002). Along this stretch of the coast, species that normally spawn offshore or nearshore can also spawn inshore, within the IRL. These species include red drum (Sciaenops ocellatus), black drum (Pogonias cromis) (Mok and Gilmore 1983; Reyier et al. 2008; R.G. Gilmore, pers. Comm. Estuarine, Coastal and Ocean Science, Inc.) and the three species that are the focus in this study: sheepshead (Archosargus probatocephalus Walbaum 1792), western Atlantic sea bream (hereafter referred to as sea bream) (A. rhomboidalis Linnaeus 1758), and pinfish (Lagodon rhomboides Linnaeus 1766), members of the family Sparidae (porgies) in the western central North Atlantic that includes 6 genera and 19 species (Powell and Greene 2002).
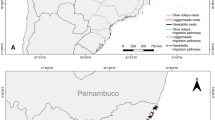
Map showing study locations of sheepshead (Archosargus probatocephalus), western Atlantic sea bream (A. rhomboidalis), and pinfish (Lagodon rhomboides). Sampling conducted by the Fisheries-Independent Monitoring program of the Fish and Wildlife Research Institute. Years indicate the initiation of sampling at each location. If sampling has been discontinued, the last year of sampling is also provided. There was no standardized FIM work for long term periods in Biscayne Bay
The sheepshead occurs in estuarine and coastal waters from Nova Scotia to Brazil, is common south of Cape Hatteras (North Carolina, 35° 15′ 2″ N/75° 31′ 43″ W) and is absent from the Caribbean Islands (Caldwell 1965; Gilhen et al. 1976; Pattillo et al. 1997). The sea bream occurs in estuarine to coastal waters from New Jersey to Argentina (Chavance et al. 1984). In the western central North Atlantic, the sea bream is common in the southern Gulf of Mexico, Central America, and the Greater Antilles, but it is rare north of the Indian River Lagoon region and, on the Gulf Coast, north of Charlotte Harbor, in southwest Florida (www.fishnet2.net, accessed 8/3/18; FWC-FWRI, Fisheries-Independent Monitoring [FIM] data; SEAMAP-SA 2000, 2017; TPW 2005). The pinfish occurs in the estuarine and coastal waters of North America from Massachusetts to Florida and Cuba and the entire Gulf of Mexico, but it is most common south of Cape Hatteras (Darcy 1985; Pattillo et al. 1997).
The sheepshead is generally known to spawn offshore (Jennings 1985), but spawning has been documented via passive acoustic surveys within the Indian River Lagoon, near habitats that support adult sea bream (R.G. Gilmore, pers. comm.). Sheepshead in spawning conditions have been found in marine waters near Hutchinson Island adjacent to the southern Indian River Lagoon, FL, primarily from January through April (Herrema et al. 1985), and young-of-the-year are most abundant in shallow estuarine areas from April through June (FWC-FWRI 2016). Sea bream can reach sexual maturity as early as five months of age (Chavance et al. 1984), and individuals in spawning condition have been collected from marine waters two to three miles from shore in the Hutchinson Island area in April, May, and September (Herrema et al. 1985). Sea bream also spawn in estuaries (Houde and Potthoff 1976; Chavance et al. 1984). Pinfish spawn primarily in coastal marine waters (Darcy 1985; Pattillo et al. 1997). They have been found in spawning conditions from November through April in the Hutchinson Island area (Herrema et al. 1985). Thus, these species overlap to some extent in habitat and reproductive timing. Additionally, these species may also overlap to some extent in spawning habitat in the Indian River Lagoon region. This situation provides ample opportunity for interactions among these species, most importantly hybridization.
Hybridization, as used here, refers to interbreeding between species in the same or a different genus. It is an occurrence during evolution that can have an adaptive or disruptive result. Hybridization is believed to be more common in fishes than in other vertebrates (Allendorf and Waples 1996), mainly because fertilization is almost exclusively external and occurs because of promoting factors such as closeness of genetic relationships, incomplete behavioral isolation, and overlapping spawning grounds and seasons. It may also be triggered by habitat changes resulting from environmental factors (Montanari et al. 2012). Anthropogenic interference can increase the rate of hybridization (Allendorf et al. 2001), but such induced hybridization may diminish the survival of the parental species (Rhymer and Simberloff 1996) or allow them to adapt to different or changing environments (Baskett and Gomulkiewicz 2011).
Putative hybrids were first collected in the IRL in 2005, during standard stratified-random sampling of the northern Indian River Lagoon (NIRL, Fig. 1). The putative hybrids were noticed among samples primarily by pigment patterns and are recognized as a form that is related, to but not identical to, pinfish, sheepshead, or sea bream (Fig. 2). The hybrids vary somewhat in pigment pattern but are generally darker than the three known species and possess elements of the pigment patterns of all three. They may have five to seven dark bars, the same as in sheepshead but different from pinfish, which has five or six relatively weak bars and relatively thicker golden and silver-blue stripes. The shoulder spot is often prominent and centered below the lateral line as in sheepshead but different from those in pinfish where it is located on the lateral line. Based on these color patterns and related external features observed in the field at the time of collections, these specimens were temporarily classified as Archosargus spp. and excluded from subsequent sheepshead stock assessments and related analyses focusing on sheepshead. At the time, the sea bream was not considered a likely parent species because it was not abundant in the core system of the NIRL (Fig. 1). Extensive phylogenetic studies of the Sparidae have been done using mtDNA markers (Chiba et al. 2009; Orrell and Carpenter 2004; Orrell et al. 2002), but none included all three species together, and the genetic relationships among the three species is unknown. In this study, to determine the genetic relationships among the three sparid species we used mitochondrial DNA regions. Additionally, we used nuclear DNA introns that are useful in the construction of phylogenetic trees (Seyoum et al. 2013; Creer 2007; Yu et al. 2011).
The objectives of this study were to (1) determine the genetic relationships among the three sparids; (2) genetically identify any hybrids and their parental types; (3) characterize the status of the hybrids by determining the microsatellite genotypic proportions of the parental types, and (4) determine the direction of hybridization. Hybrids and the parental types can be identified by several techniques, of which microsatellite markers have significant power to detect different hybrid statuses (Anderson and Thompson 2002). To identify and characterize the status of the hybrids and to identify the parental types we used 18 microsatellite markers selected from sheepshead, as well as from other sparids (Seyoum et al. 2016). To determine the maternal parent of the hybrids we sequenced the mtDNA 16S rRNA of all the individuals that were genetically identified as hybrids. The mtDNA is maternally inherited and provides the means to identify the maternal parent of the hybrids.
Materials and methods
Sample collection and DNA extraction
The FIM program uses a stratified random sampling design to monitor nekton health, species diversity, abundance, and other parameters in all of Florida’s major estuarine systems (Fig. 1). This method involves the use of multiple types of gear, including a 183 m × 3 m, center-bag haul seine (38 mm stretched mesh) to representatively sample large-bodied fishes along shoreline habitats in waters < 2.5 m deep. The standardized dimensions of the area sampled by this seine are approximately 40 × 103 m, or 4120 m2. Based on geographic criteria and sampling logistics, each estuary has been divided into large zones that define areas of biological and hydrological homogeneity. Zones are divided into a grid of 1.0 nm × 1.0 nm cartographic cells (nm = nautical mile), each of which is further divided into a microgrid of 0.1 nm × 0.1 nm cells.
All fish captured from the Northern Indian River Lagoon (NIRL) and the Southern Indian River Lagoon (SIRL) (Fig. 1), were identified in the field to the lowest practical taxon, measured for standard length (SL; mm), and counted using standardized procedures (FWC-FWRI 2016). During 2015, we collected representative samples of 172 sea bream (NIRL, N = 52; SIRL, N = 120 and 232 pinfish (NIRL, N = 47; SIRL, N = 185) (Fig. 1). Also, we received 36 specimens that were initially identified as Archosargus spp, collected and retained over a few years for genetic identification. Furthermore, in this study, we included 47 sea bream collected from Maracaibo, Venezuela, in 1999. From each fish sampled, a small piece of dorsal fin was excised and placed in 70% ethanol. Total DNA was extracted using the PureGene DNA isolation kit (Gentra Systems Inc., Minneapolis, MN) and was rehydrated in 50 µl of deionized water. Sheepshead specimens collected in a previous study from Sebastian Inlet, Florida, to South Carolina (N = 157, Seyoum et al. 2017), assumed to be nonhybrids, were used as reference specimens for hybrid analysis.
Genetic relationship
To investigate the genetic relationships among the three sparids, we conducted DNA sequencing for eight to 17 decidedly identified individuals from each taxon. Sequencing was completed from three partial regions of the mitochondrial DNA [16S rRNA; COI (cytochrome oxidase subunit 1) and cyt b (cytochrome b)] and three Exon-primed intron-crossing (EPIC) nDNA markers [EPIC1 (hypothetical protein gene), EPIC2 (proliferator-activated receptor gamma protein gene), and EPIC7 (spectrin alpha 2 gene)] (Table 1). To determine the species of the maternal parent, we sequenced the16S rRNA of all the hybrids that were verified by genetic identification.
The polymerase chain reaction (PCR) was conducted in an Eppendorf Master Cycler Pro Series thermal cycler, using a touch-down protocol described in Seyoum et al. (2016). Briefly, the PCR product was gel-purified (Agilent Technologies, Santa Clara, CA) and cycle-sequenced from both directions using Big Dye™ Terminator Cycle-Sequencing Ready-Reactions with AmpliTaq FS DNA polymerase (Applied Biosystems, Inc., Foster City, CA). The cycle sequencing product was then precipitated with ethanol and resuspended in 22 μl of HiDi formamide and visualized on an ABI 3130 XL genetic analyzer (Applied Biosystems). The sequences obtained from each species were aligned and edited using Sequencher (v4.9, Gene Codes Corporation, Ann Arbor, MI).
Phylogenetic analysis
We evaluated the sequences of the three sparid species for genetic relationships using three methods of analysis: maximum likelihood (MEGA, v7.0; Tamura et al. 2013), Bayesian (MrBayes, v3.0; Ronquist and Huelsenbeck 2003), and maximum parsimony (PAUP* v4.0; Swofford 2000). Two analyses were conducted with the aligned sequences where (1) the DNA sequences from each gene were analyzed independently and (2) all the sequences from one genome (nuclear or mtDNA) were analyzed together by concatenating the sequences. The optimal model of sequence evolution for each partition sequence was assessed with the maximum likelihood method and Akaike information criterion procedures implemented in MEGA. Bayesian analysis was conducted by running MrBayes for ten metropolis-coupled Markov chain Monte Carlo generations, and trees were sampled every 100 generations during the run. In the concatenated sequence analyses, mixed models were used where each partition had its sequence evolution model. Parsimony analysis was performed under the alltrees search option, with which the relative clade support was assessed with 1000 bootstraps. For comparative purposes, specimens of knobbed porgy (Calamus nodosus), the closest relative to the above species (Orrell et al. 2002; Orrell and Carpenter 2004), were also sequenced except for COI and cytb, for which data were extracted from GenBank (Table S1). To simulate a better phylogenetic analysis with the data we have for the four sparid species, we extracted sequences of eight other closely related sparid species from GenBank and aligned them with our sequences using Sequencher. This simulation of the phylogenetic analysis was done only for the mtDNA data since there is no comprehensive entry for the nDNA markers for other sparids.
Microsatellite genotyping data and analysis
All specimens of sea bream, putative hybrids, sheepshead, and pinfish collected were genotyped for 12 sheepshead-specific microsatellite markers (Apro-markers; Seyoum et al. 2016) and six sparid markers (Table S2). Multiplex PCR amplification of each specimen was carried out with three pairs of primers, with each forward primer labeled with a unique fluorescent dye as described in Seyoum et al. (2016). Fragments were visualized on an ABI 3130 XL genetic analyzer using the Gene Scan-500 ROX-labeled size standard and genotyped using GENEMAPPER software v4.0 (Applied Biosystems).
Tests for linkage disequilibrium, Hardy–Weinberg equilibrium (HWE), and observed (HO) and expected (HE) heterozygosity estimates (with Bonferroni corrections) were conducted using GENEPOP (v3.4, Rousset 2008). Estimates of the number of alleles, polymorphic information content, and frequencies of null alleles were calculated using programs implemented in CERVUS (v3.0.7; Kalinowski et al. 2007).
Bayesian clustering analysis was conducted using STRUCTURE (version 2.3.4, Pritchard et al. 2000) to verify the number of species we had in our data set. Parameters consisted of 10 replicate simulations using 2.0 × 105 Markov–Chain Monte Carlo repetitions after a 1.0 × 106 burn-in period for each value of K (1–6), with the admixture model and independent-allele-frequencies option. To visualize the output for the ten replicated runs from STRUCTURE, we used STRUCTURE HARVESTER (version 0.56.3; Earl and von Holdt 2012) which uses the posterior probabilities from STRUCTURE to determine the optimal K value according to the simulation method of Evanno et al. (2005). The optimal K value is expected to be equal to the number of genetic groups.
Genetic identification of hybrids
We evaluated the existence of hybrids, that is, individuals with admixed genotypic proportions from two species, with three methods:
-
(1)
From posterior probabilities of microsatellite genotypic proportion assignments (q-values) determined from the Bayesian clustering analysis for K = 3, we assessed the identification of hybrids and the determination of their parentage. Q-values are obtained from the ten replicated runs at K = 3 compiled by STRUCTURE HARVESTER and further aligned and summarized using the program CLUMPP (Jakobsson and Rosenberg 2007).
-
(2)
To explore the interspecific variance between sea bream, sheepshead and their hybrids we conducted a discriminant analysis of principal components (DAPC: Jombart et al. 2010) in the R package Adegenet (Jombart 2008). The plot enables visualization of the position and distribution of hybrids relative to those of the parental individuals. Introgressive hybridization would be indicated by a continuous distribution of the hybrid aggregates abutting that of both parentals. The species that did not contribute to hybridization was excluded from the DAPC analysis.
-
(3)
To determine the statistical power of hybrid detection and to classify hybrids into different generations, we used the program HYBRIDLAB (Nielsen et al. 2006; Schwartz and Beheregaray 2008; Alvarez et al. 2015), which estimates allele frequencies from user-specified parental populations from either actual genotyped populations or simulated populations. The estimated allele frequencies are then used to create multilocus genotypes for user-specified populations. In this study, genotypes were simulated for the identified hybrid parental populations, each with 100 selected individuals that showed > 98% genotypic proportion (q-values) estimated in the STRUCTURE runs. These selected individuals are used to build up simulated pure parental populations that were then used to simulate F1 and F2 hybrids, first-generation backcrosses (BC) and second-generation (double) backcrosses (DBC) to each of the parental populations. After completing the simulations for sheepshead and sea bream, the genotypes from each cross were evaluated in STRUCTURE using the admixture and independent allele frequencies model at K = 2. The genotypic proportion assignments were first arcsine-transformed to stabilizes the variance and to normalize the proportional data; 95% and 99% confidence intervals were determined to estimate the minimum thresholds (q-values) for pure and hybrid individuals as in Litrell et al. (2007).
Results
Phylogenetic analysis
Overall, the three methods of the phylogenetic analyses, maximum likelihood, Bayesian, and parsimony, produced identical topologies in all analyses with slight differences in bootstrap values. The results based on the concatenated sequences of the mtDNA and nuclear intron DNA showed soft polytomy with sea bream and sheepshead as sister species (Fig. 3). This relationship was not consistently shown by each of the three mtDNA regions and the three nuclear DNA introns. Instead, markers within the mtDNA and nuclear DNA introns showed unresolved and alternate possibilities of genetic relationships between sea bream and sheepshead and pinfish and sheepshead (Figs. S1, S2). The genetic distances from the concatenated mtDNA and the nuclear DNA sequences were virtually the same between sea bream and sheepshead as between sheepshead and pinfish (Table 1) but were inconsistent from each of the markers (Table S3). There were no amino acid differences in COI but in 368 amino acids in cyt b there were three differences between pinfish and sheepshead, ten between sheepshead and sea bream and seven between pinfish and sea bream (Table S3). More genetic information would be needed to decidedly resolve the relationship between the three species.
Phylogenetic relationships of four sparid species (Archosargus probatocephalus, A. rhomboidalis, Lagodon rhomboides, and Calamus nodosus) based on the concatenated sequences of three mtDNA regions constructed with eight other closely related sparids extracted from Orrell and Carpenter (2004); and three concatenated nDNA introns. Bootstrap values (1000 replicates) are shown on the branches in % for Maximum Likelihood/MrBayes/Maximum Parsimony. Bootstrap values of only maximum likelihood are given at nodes of the extracted eight species in the mtDNA topology. Substitution models based on the Akaike information criterion procedures in MEGA are given in Table 1. Crescent shapes at terminal nodes of the nDNA topology indicate collapsed branches of four or more (median eight) individuals varying between one and three base pairs
Microsatellite markers
Sheepshead (N = 157), sea bream (N = 257), and pinfish (N = 232) were assayed at 18 microsatellite markers (12 selected from sheepshead and six from other sparids). Sea bream from IRL showed significantly less gene diversity and fewer alleles per locus (0.3912, 6.9) than did sea bream from Venezuela (0.5071; 8.2), pinfish (0.7995; 20.3), or sheepshead (0.7238; 14.5) (Table S2). The overall number of alleles observed and the heterozygosity and polymorphic information content of sea bream (IRL) were significantly lower than those for the sea bream from Venezuela and the sheepshead and pinfish from IRL (Table S2). Visualization of the STRUCTURE analysis revealed a single optimal peak at K = 3 (∆ K = 1190) confirming that there were only three genetic groups in our data set.
Genetic identification of hybrids
Hybrids were identified based on 18 multilocus microsatellite genotypes and 10 replicate STRUCTURE runs at K = 3. Thirty-six specimens that were morphologically identified as hybrids and two other suspected hybrids aligned in the cluster of the reference specimens identified as sea bream collected from IRL (Fig. 4a). These individuals with two different genotypic contributions are numbered one to 36 for identification purposes, with the mean genotypic proportions of every individual given in Table 2 and, graphically shown in Fig. 4b.
a Genetic clustering of Archosargus probatocephalus (sheepshead), A. rhomboidalis (western Atlantic sea bream), and Lagodon rhomboides (pinfish) based on individual assignment probabilities (q-values) from STRUCTURE at the optimal K value of 3, where K is the number of genetic groups. If the individual has genotypic proportion assignments of more than one parent species, the vertical bar may be partitioned into segments, in which the vertical height of each shade represents the genotypic proportion of each species: b bar graph of the partial enlargement of the STRUCTURE analysis encompassing the region containing the macroscopically identified 36 hybrids and the suspected introgressive hybrids identified as X and Y. The genotypic proportion of each parental species is given in Table 2
The visual scattergram of the interspecific discriminant analysis of principal components (DAPC) showed three clusters, that of the two parental types sheepshead and sea bream and the 36 morphologically and genetically identified hybrids (Fig. 5). The hybrid cluster is defined separately but its position was not exactly between those of the parental clusters. It is shifted towards the sheepshead cluster because 23 of the 36 hybrids had significantly greater sheepshead genotypic proportions relative to sea bream while in the remaining 13 the proportions are about equal. The two individuals X and Y with significantly less sheepshead genotypic proportion (Fig. 4a, b; Table 2) were not in the hybrid cluster but were subsumed in the sea bream cluster.
Discriminant analysis of principal components (DAPC) clustering of sheepshead (N = 157) and western Atlantic sea bream (N = 210), based on 18 microsatellite loci. All 36 individuals morphologically and genetically identified as hybrids (Table 2) were subsumed into the hybrid cluster whereas individuals X and Y were subsumed in the sea bream cluster. Because pinfish did not contribute to hybridization it was excluded from the DAPC analysis
Threshold assignment values
The distributions of assignment values of q for five simulated classes of hybrids based on the simulation of 200 individual genotypes in HYBRIDLAB and the admixture algorithm implemented in STRUCTURE for K = 2 are plotted in Fig. S3. The mean genotypic proportion assignments to sheepshead and sea bream genetic clusters (q ± SD) for simulated and different classes of hybrids are given in Table S4. These distributions showed a significant overlap between adjacent hybrid classes. The classification of the parental genotypic proportion determined from ten replicate runs in STRUCTURE at K = 3 identified 36 well-defined hybrids and two more less-defined apparent hybrids identified as X and Y (Fig. 4; pinfish did not have any contribution). None of the hybrids had equal genotypic proportions because the microsatellite loci are not all fixed and so are not species-diagnostic.
The 36 individuals that were morphologically identified as hybrids have significant admixed genotypic proportions (Table 2; Fig. 4b). Twenty-five of these were decidedly F1 hybrids at 95% confidence interval (CI). At 99% CI, however, all 36 fell within the F1 hybrid thresholds. The range of values was larger for the higher CI due to the expectation that it would capture a greater proportion of F1 hybrid individuals. The maternal parent species of all the 36 hybrids based on the mtDNA16S rRNA sequence, was sheepshead, but not for the two individuals that are less-defined as hybrids (X and Y), which were sea bream. Individuals X and Y initially listed as sea bream during macroscopic identification at the time of collection were sequenced for the EPIC1 and EPIC7 loci and found to be homozygous for both. Furthermore, they were also subsumed in the sea bream cluster in the DAPC analysis. For these reasons, the genotypic proportions of individuals X and Y may have been the result of microsatellite marker artifacts due to interference with genotyping (Olejniczak and Krzyzosiak 2006). If they were double backcrosses, however, the frequency of introgressive hybridization between sea bream and sheepshead would be estimated at 1%. This implies that hybridization between sea bream and sheepshead would be highly asymmetrical, that is, sea bream males mate with sheepshead females (Table 2). Three other individuals, #1, #5, and #36 which were also suspected as backcrosses were subsumed in the hybrid cluster. Their nDNA sequences also had hybrid genotypes. The absence of abundant introgressive hybrids and the significant genetic divergence between sea bream and sheepshead suggests that their F1 hybrids may be sterile to some extent. Any prediction of large-scale hybrid sterility in this study is not warranted. Additional research would be needed to determine the degree of hybrid sterility between these two species.
The 36 hybrid specimens initially listed as Archosargus spp. were part of 69 hybrids retained for genetic identification. Unfortunately, the first 33 were recorded but not retained for genetic study. The 69 morphologically identified as hybrids were recorded by FIM out of 678 sea bream collected over 11 years (2006–2016). Therefore, the most reliable estimate of hybridization between sheepshead and sea bream is 10.2%, which indicates a significant interaction between the two species.
Discussion
Our intention to study the phylogenetic relationships among the three sparid species was inspired by frequent observations of specimens that appeared to be hybrids in the IRL since 2005. These specimens cataloged as Archosargus spp. appeared to exhibit external characteristics intermediate between those of the intergeneric species sheepshead and pinfish, rather than between the congeners sheepshead and western Atlantic sea bream. Phylogenetic relationships among the three sparid species based on the combined sequence data of three mtDNA regions and three nDNA introns indicated soft polytomy, with sea bream and sheepshead as sister species. Analysis of genetic divergence showed that there are significant divergences between the three species. The genetic divergence between sea bream and sheepshead and between pinfish and sheepshead is virtually equal. The divergence between sea bream and pinfish, however, is greater than that of the divergence between each of them to sheepshead but no pinfish-sheepshead hybrids have been recorded.
The spawning seasons of sheepshead and pinfish overlap in the IRL region (Herrema et al.1985), and both species generally tend to spawn outside of estuaries (Darcy 1985; Jennings 1985; Render and Wilson 1992; Pattillo et al. 1997). Furthermore, young-of-the-year of both sheepshead and pinfish have often been collected at the same time in the IRL during recruitment periods (FIM, unpublished data), suggesting that the spawning areas, larval sources, and oceanographic influences for these two species are similar. Thus, hybridization between sheepshead and pinfish is possible, but no such hybrids were apparent. At least part of the IRL population of sheepshead might spawn in the lagoon (R.G. Gilmore pers. comm.), or outside the lagoon, or the two species might spawn in different areas, perhaps even at different depths. In the absence of a directed search for hybrids, sheepshead–pinfish hybrids may escape detection because their markings may be more difficult to distinguish than those of sheepshead-sea bream hybrids that are easily noticed. Or sheepshead-pinfish may altogether have lower viability. Further directed search for sheepshead–pinfish hybrids would be needed to investigate these possibilities. Pinfish and sea bream, on the other hand, may be less likely to hybridize because they are more different in life-history parameters and genetic divergence, as shown in this study. But hybridization between these species under a drastically altered environment cannot be ruled out.
All hybrids identified in this study were between sea bream and sheepshead. This finding may seem counterintuitive at first glance because the sea bream is generally thought to spawn within estuaries (Houde and Potthoff 1976; Chavance et al. 1984), while the sheepshead is generally thought to spawn outside of estuaries. If some sheepshead in the IRL spawns in the lagoon, however, then the spawning areas of these two species could overlap. Inshore spawning in the IRL by normally offshore-spawning species is well documented, with red drum and black drum being prominent examples (Mok and Gilmore 1983; Reyier et al. 2008; R.G. Gilmore pers. comm.).
Three results characterize the hybridization between sheepshead and sea bream: (1) it is asymmetric (based on 16S rRNA, sheepshead is the maternal contributor to the hybrids); (2) the hybridization frequency is high (10.2%), and (3) the hybrids are nearly all F1.
Asymmetric hybridization may be related to a bias toward the more abundant of the two species, sheepshead, which is approximately 38 times more abundant than sea bream in the NIRL and approximately four times as abundant in the SIRL (FIM unpublished data). Such asymmetric hybridization based on abundance has been observed between the brown trout (Salmo trutta) and Atlantic salmon (S. salar) (Álvarez and Garcia-Vazquez 2011).
Because sheepshead–sea bream F1 hybrids can be easily identified based on macroscopic examination, FIM’s 11-year record of 10.2% frequency of hybrids is considered reliable. If there had been any introgressive hybrids, they may not have been easy to visually identify and could have been missed. For example, individuals, X, and Y which were probably introgressive hybrids were not visually identified as hybrids. Nevertheless, the examination of the hybrid karyotype is the best method for determining the degree of hybrid sterility which has yet to be conducted.
The high frequency of sheepshead–sea bream hybridization indicates that a larger proportion of hybrids are produced relative to the smaller population size of sea bream than to the larger population size of sheepshead. This terminal hybridization does not entail a genetic issue but certainly becomes an ecological and demographic burden, particularly if the hybrids are effective competitors that could negatively affect one or both parental species and other fish species in the community through invasion and competition for food and space. Possible results include loss of reproductive potential, suppression of population growth rates needed for biomass replacement, and increased risk of genomic extirpation of the parental species (Senanan et al. 2004; Allendorf et al. 2013). A simulation study showed that from competition alone, hybridization increases the risk of extinction of species (Wolf et al. 2001). An excellent example of a destructive outcome of terminal hybridization is seen in cyprinid fishes in Japan, between shinai-motsugo (Pseudorasbora pumila) females and motsugo (P. parva) males. This asymmetric hybridization is leading to the rapid replacement of P. pumila by P. parva (Konishi and Takata 2004) to the extent that P. pumila is now classified as an endangered species (IUCN RDB category) by the Ministry of the Environment of Japan (1997). In a laboratory experiment, it was shown that male P. pumila also hybridize with female P. parva, producing sterile hybrids (paper in Japanese, cited by Konishi and Takata 2004). This reciprocal hybridization, however, was not observed in nature. Sheepshead males may also hybridize with sea bream females but were not observed because the cross-fertilization may fail to develop. Hence, there could even be a higher loss of hidden reproductive potential for sea bream because of the possibility of nonviable reciprocal hybridization, particularly if there is mate pairing. However, there are no recorded observations of mate pairing in the three sparid fishes. If the rate of terminal hybridization between sea bream and sheepshead increased, the eventual fate of sea bream in the IRL could be extirpation.
The sheepshead has been documented as an abundant species in the IRL since the 1800s (Wilcox 1897). On the other hand, the sea bream was known to be common in the Florida Keys–Florida Bay–Biscayne Bay region from the 1800s through recent years (Jordan 1884; Hammerschlag and Serafy 2010; www.fishnet2.net, accessed Aug 8, 2018) but apparently did not move north into the IRL until the 1960s (Christensen 1965; Gilmore 1977) and may not have been common there until the 1990s (FIM unpublished data, this study). Conditions seemed perfect for hybridization with sheepshead as early as 1995, after sea bream expanded northward and encroached on spawning habitat and overlapping spawning seasons with sheepshead, but hybrids were not seen until 2005. The question is why? We believe that the distinctive appearance of the hybrids (especially in terms of pigment and pattern) makes it very unlikely that biologists would not have noticed any hybrids collected in the intensively surveyed IRL, making the 10-year delay in collecting them even more unusual. In southeast Florida, including Biscayne Bay, the Keys, and eastern Florida Bay, the sea bream is also more populous than is the sheepshead in most estuaries (Flaherty et al. 2013; Hammerschlag and Serafy 2010), but no hybrids have been reported there. Within the western North Atlantic, sheepshead are found alone in estuaries north of central Florida (FIM unpublished data; Akin et al. 2003; Stunz et al. 2010), sea bream are found alone in areas such as Cuba and Venezuela (Claro et al. 2001; De Grado and Bashirullah 2001), and the two species occur together, without known hybridization, in estuaries in the southern Gulf of Mexico (Ramos-Miranda et al. 2005; Palacios-Sánchez et al. 2015). In the latter estuaries, sea bream is the distinct numerical dominant. The absence of any report of hybridization between these species could be because the fish were not expected to hybridize and hence no search was made.
The onset of considerable hybridization between sheepshead and sea bream in the IRL, perhaps as much as a decade after sea bream began to be recorded there in large numbers, and no report of hybridization anywhere else makes this occurrence unique. It appears that this hybridization may have been triggered by increasing anthropogenic stressors in the IRL. This term encompasses multiple factors that could harm the ecosystem and has been connected to the breakdown of mechanisms that affect reproductive isolation between species, facilitating hybridization (Rhymer and Simberloff 1996; Mullen et al. 2012; Crego-Prieto et al. 2012). For decades, the IRL ecosystem has been a focus of research on human activities that harm ecological processes and threaten dire consequences for fish populations. The results of those activities include declining water quality, harmful algal blooms, pollution, habitat degradation, and loss, and bioaccumulation of harmful chemical compounds (Johnson-Restrepo et al. 2005; Xue et al. 2017; Barile 2018; Adams et al. 2019). The IRL has been undergoing chronic, anthropogenic ecological disturbances, which could break down isolation mechanisms and facilitate hybridization between fish that do not normally hybridize because they are quite divergent, as this study showed. Further research is needed to determine the relative contributions of the sea bream’s range expansion, the unique geomorphological characteristics of the IRL, and the many types of anthropogenic stressors (and their means of action) in eroding isolation mechanisms between sea bream and sheepshead in the IRL.
The sheepshead–sea bream hybridization in the IRL may be a small part of a larger occurrence of hybridization in places where the species already share spawning habitat and where anthropogenic stressors are prevalent. Since the pinfish has a similar divergence from the sheepshead as the sea bream, hybridization between sheepshead and pinfish, too, might be facilitated by disturbances of the natural environmental conditions. A hint that hybridization occurs between sparids other than between sheepshead and sea bream comes from an individual identified as pinfish (L. rhomboides) from Bahia Honda in the Florida Keys in Orrell and Carpenter’s (2004) construction of a phylogeny of the Sparidae. This individual had the mtDNA 16S rRNA gene sequence of the sea bream, not that of pinfish. Thus, it could be a hybrid between a male pinfish and a female sea bream, signifying hybridization between these highly divergent species. However, it could also have been a case of misidentification. Environmental disturbance is not limited to the IRL. Florida Bay is also affected by multiple stressors (Glibert et al. 2009) and a similar occurrence of hybridization as in the IRL could be expected to occur in this Bay including Biscayne Bay. Unfortunately, FIM ceased surveying in Florida Bay in 1997 and the Florida Keys in 2004, just before the first sheepshead–sea bream hybrids were observed in the IRL, in 2005. There has been no long-term monitoring by FIM in Biscayne Bay other than short term intermittent fisheries work. Hence, in the absence of any study, the occurrences of hybridization or hybrids in these Bays are unknown.
Although fishery managers had no reason to suspect hybridization among the three sparids, these activities may have been occurring unnoticed since 2005 in some places, presumably induced by increasing anthropogenic stressors. Based on this study, hybridization among the three species would most likely produce terminal hybrids that could, on a larger scale, lead to significant negative effects on the population dynamics involving parental species and other community members (Senanan et al. 2004; Allendorf et al. 2013). Therefore, hybridization among the three species and the assessment of whether it is accelerating in response to increasing anthropogenic interference in the IRL needs to be determined. Periodic estimates of the extent of hybridization between sheepshead and sea bream could (1) be used to monitor the risk of extirpation or depletion of sea bream, the species most likely to be affected and (2) serve as a sensitive indicator of anthropogenic interference and the overall IRL health.
Sheepshead–sea bream hybridization should have only minimal effects on fishery-independent data for stock assessments of sheepshead in the IRL since the hybrids can be easily identified visually at adult sizes. But if juveniles were to be included in population dynamics studies, evaluating the effect of hybridization would be more difficult because coloration patterns and other identifying characteristics of hybrids may not yet be evident in small juveniles, complicating, for example, assessments of recruitment and relative abundance of juveniles. Another concern, even in adult fish, is that stock assessments rely on fishery-dependent data (i.e., reports of landings by recreational and commercial fishers), where sheepshead–sea bream hybrids could be misreported as sheepshead in the absence of any instruction to avoid this misidentification. In Florida, sheepshead-specific regulations promote the effective conservation of that species. But while pinfish and sea bream are monitored in Florida waters, both are classified as unregulated species. The results of this study will help guide future management decisions for pinfish and sea bream in the region. Additional research is needed to improve our understanding of the presence, causes, and effects of hybridization and its effect on the population dynamics among the three species.
Data availability
The accession numbers of the mtDNA region and nDNA locus sequences of the sparid species deposited in GenBank are given in Table S1.
References
Adams DH, Tremain DM, Paperno R, Sonne C (2019) Florida lagoon at risk of ecosystem collapse. Science 365:991–992
Akin S, Winemiller KO, Gelwick FP (2003) Seasonal and spatial variations in fish and macrocrustacean assemblage structure in Mad Island Marsh estuary, Texas. Estuar Coast Shelf Sci 57(1–2):269–282
Allendorf FW, Leary RF, Spruell P, Wenburg JK (2001) The problems with hybrids: setting conservation guidelines. Trends Ecol Evol 16:613–622
Allendorf FW, Luikart G, Aitken SN (2013) Hybridization. Conservation and the genetics of populations, 2nd edn. Wiley, West Sussex, pp 352–376
Allendorf FW, Waples RS (1996) Conservation and genetics of salmonid fishes. In: Avise JC, Hamrick JL (eds) Conservation genetics: case histories from nature. Chapman and Hall, New York, pp 238–280
Alvarez AC, Peterson D, Taylor TA, Tringali MD, Barthel BL (2015) Distribution and amount of hybridization between shoal bass and the invasive spotted bass in the lower Flint River, Georgia. Am Fish Soc Symp 82:503–521
Álvarez D, Garcia-Vazquez E (2011) Maintenance of asymmetric hybridization between Atlantic salmon (Salmo salar) and brown trout (Salmo trutta) via postzygotic barriers and paternal effects. Can J Fish Aquat Sci. 68:593–602
Anderson EC, Thompson EA (2002) A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160:217–1229
Barile PJ (2018) Widespread sewage pollution of the Indian River Lagoon system, Florida (USA) resolved by spatial analyses of macroalgal biogeochemistry. Mar Pollut Bull 128:557–574
Baskett LM, Gomulkiewicz R (2011) Introgressive hybridization as a mechanism for species rescue. Theor Ecol 4:223–239
Caldwell DK (1965) Systematics and variation in the sparid fish Archosargus probatocephalus. Bull South Calif Acad Sci 64:89–100
Chavance P, Flores-Coto C, Sanchez-Iturbe A (1984) Early life history and adult biomass of sea bream in the Terminos Lagoon, southern Gulf of Mexico. Trans Am Fish Soc 113:166–177
Chiba SN, Iwatsuki Y, Yoshino T, Hanzawa N (2009) Comprehensive phylogeny of the family Sparidae (Perciformes: Teleostei) inferred from mitochondrial gene analyses. Genes Genet Syst 84:153–170
Christensen RF (1965) An ichthyological survey of Jupiter Inlet and Loxahatchee River, Florida. M.S. thesis, Florida State University
Claro R, Lindeman KC, Parenti LR (2001) Ecology of the marine fishes of Cuba. Smithsonian Institution Press, Washington, DC
Creer S (2007) Choosing and using introns in molecular phylogenetics. Evol Bioinform 3:99–108
Crego-Prieto V, Martinez JL, Roca A, Garcia-Vazquez E (2012) Interspecific hybridization increased in congeneric flatfishes after the Prestige oil spill. PLoS ONE 7(4):e34485
Darcy GH (1985) Synopsis of biological data on the pinfish, Lagodon rhomboides (Pisces: Sparidae). Technical report NMFS 23. NOAA, Springfield VA
De Grado AA, Bashirullah A (2001) Some attributes of community structure of fishes in Laguna Grande de Obispo, Golfo Cariaco, Sucre State, Venezuela. Acta Cient Venez 52:3–13
Dybas CL (2002) Florida’s Indian River Lagoon: an estuary in transition. Bioscience 52:554–559
Earl DA, von Holdt MB (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Flaherty KE, Matheson RE Jr, McMichael Robert H Jr, Perry WB (2013) The influence of freshwater on nekton community structure in hydrologically distinct basins in northeastern Florida Bay, FL, USA. Estuar Coast 36:918–939
FWC-FWRI (Florida Fish and Wildlife Conservation Commission, Fish and Wildlife Research Institute) (2016) Fisheries-Independent Monitoring program 2016 annual report data summary report. In-house Report IHR2017-003. FWRI, St. Petersburg, FL
Gilhen J, Gruchy CJ, McAllister DE (1976) Sheepshead, Archosargus probatocephalus, and the feather blenny, Hypsoblennius hentzi, two additions to the Canadian arctic ichthyofauna. Can Field Nat 90:42–46
Gilmore RG Jr (1977) Fishes of the Indian River Lagoon and adjacent waters, Florida. Bull Fla State Mus Biol Sci 22:101–148
Gilmore RG Jr (1995) Environmental and biogeographic factors influencing ichthyofaunal diversity: Indian River Lagoon. Bull Mar Sci 57:153–170
Glibert PM, Heil CA, Madden CJ (2009) Florida Bay: a subtropical system increasingly influenced by multiple stressors. Contrib Mar Sci 38:1–4
Hammerschlag N, Serafy JE (2010) Nocturnal fish utilization of a subtropical mangrove–seagrass ecotone. Mar Ecol 31:364–374
Herrema DJ, Peery BD, Williams-Walls N, Wilcox J (1985) Spawning periods of common inshore fishes on the Florida east coast. Northeast Gulf Sci 7:153–155
Houde DD, Potthoff T (1976) Egg and larval development of the sea bream Archosargus rhomboidalis. Bull Mar Sci 26:506–529
Jakobsson M, Rosenberg N (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jennings CA (1985) Species profiles: 1ife histories and environmental requirements of coastal fishes and invertebrates (Gulf of Mexico): sheepshead. U.S. Fish Wildl. Serv. Biol. Rep. 82 (11.29). U.S. Army Corps of Engineers, TR EL-82-4 10 pp
Johnson-Restrepo B, Kannan K, Addink R, Adams DH (2005) Polybrominated diphenyl ethers and polychlorinated biphenyls in a marine food web of coastal Florida. Environ Sci Technol 39:8243–8250
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jordan DS (1884) The fishes of the Florida Keys. Bull United States Fish Comm 4:77–80
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106
Konishi M, Takata K (2004) Impact of asymmetrical hybridization followed by sterile F1 hybrids on species replacement in Pseudorasbora. Conserv Genet 4:463–474
Litrell BM, Lutz-Carrillo DJ, Bonner TH, Fries LT (2007) Status of an introgressed guadalupe bass population in a Central Texas stream. N Am J fish Manage 27:785–791
Ministry of the Environment of Japan (1997) Red list of threatened fishes of Japan. Japan Integrated Biodiversity Information system, Tokyo. https://www.biodic.go.jp/english/J-IBIS.html. Accessed 4 Oct 2018
Mok H-K, Gilmore RG Jr (1983) Analysis of sound production in estuarine aggregations of Pogonias cromis, Bairdiella chrysoura, and Cynoscion nebulosus (Sciaenidae). Bull Inst Zool Acad Sinica 22:157–186
Montanari SR, Van Herwerden L, Pratchett MS, Hobbs J-PA, Fugedi A (2012) Reef fish hybridization: lessons learnt from butterflyfishes (genus Chaetodon). Ecol Evol 2:310–328
Mullen SP, Little K, Draud M, Brozek J, Itzkowitz M (2012) Hybridization among Caribbean damselfish species correlates with habitat degradation. J Exp Mar Biol Ecol 416–417:221–229
Nielsen EE, Bach LA, Kotlicki P (2006) HYBRIDLAB (v1.0): a program for generating simulated hybrids from population samples. Mol Ecol Notes 6:971–973
Olejniczak M, Krzyzosiak WJ (2006) Genotyping of simple sequence repeats—factors implicated in shadow band generation revisited. Electrophoresis 27:3724–3734
Orrell TM, Carpenter KE (2004) A phylogeny of the fish family Sparidae (porgies) inferred from mitochondrial sequence data. Mol Phylogenet Evol 32:425–434
Orrell TM, Carpenter KE, Musick JA, Graves JE (2002) Phylogenetic and biogeographic analysis of the Sparidae (Perciformes: Percoidei) from cytochrome b sequences. Copeia 3:618–631
Palacios-Sánchez S, Vega-Cendajas ME, Hernández M (2015) Ichthyological survey on the Yucatan coastal corridor (southern Gulf of Mexico). Rev Biodivers Neotrop 5:145–155
Pattillo ME, Czapla TE, Nelson DM, Monaco ME (1997) Distribution and abundance of fishes and invertebrates in Gulf of Mexico estuaries. Volume II: species life history summaries. ELMR Rep. No. 11. NOAA/NOS Strategic Environmental Assessments Division, Silver Spring, MD
Powell AB, Greene MD (2002) Preliminary guide to the identification of the early life history stages of sparid fishes of the western central North Atlantic. NOAA Technical Memorandum NMFS-SEFSC-480. NOAA, Silver Spring, MD
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multi-locus genotype data. Genetics 155:945–959
Ramos-Miranda J, Quiniou L, Flores-Hernandez D, Do-Chi T, Ayala-Perez L, Sosa-Lopez A (2005) Spatial and temporal changes in the nekton of the Terminos Lagoon, Campeche, Mexico. J Fish Biol 66:513–530
Render JH, Wilson CA (1992) Reproductive biology of sheepshead in the northern Gulf of Mexico. Trans Am Fish Soc 121:757–764
Reyier EA, Shenker JM, Christian D (2008) Role of an estuarine fisheries reserve in the production and export of ichthyoplankton. Mar Ecol Prog Ser 359:249–260
Rhymer JM, Simberloff D (1996) Extinction by hybridization and introgression. Annu Rev Ecol Syst 27:83–109
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Rousset F (2008) Genepop’007: a complete reimplementation of the GenePop software for Windows and Linux. Mol Ecol Resour 8:103–106
Schwartz T, Beheregaray L (2008) Using genotype simulations and Bayesian analysis to identify individuals of hybrid origin in Australian bass: lessons for fisheries management. J Fish Biol 72:435–450
SEAMAP-SA (2000) SEAMAP-SA 10-year trawl report: results of trawling efforts in the coastal habitat of the South Atlantic Bight, FY 1990–1999. Special Tech. Rep. No. 71. Atlantic States Marine Fisheries Commission, Arlington, VA
SEAMAP-SA (2017) SEAMAP-SA 10-year trawl report: results of trawling efforts in the coastal habitat of the South Atlantic Bight, 2016. Rep. No. SEAMAP SA-CS–2016-004. South Carolina Department of Natural Resources, Columbia, SC
Senanan W, Kapuscinski AR, Na-Nakorn U, Miller LM (2004) Genetic impacts of hybrid catfish farming (Clarias macrocephalus × C. gariepinus) on native catfish populations in central Thailand. Aquaculture 235:167–184
Seyoum S, Barthel LB, Tringali MD, Carney S (2013) Phylogenetic relationships among four western Atlantic Cynoscion species based on DNA sequences from 11 nuclear introns, two mitochondrial genes, and genotypes from 32 microsatellite markers. Gulf Mex Sci 1–2:40–49
Seyoum S, McBride RS, Puchutulegui C, Dutka-Gianelli J, Alvarez AC, Panzner K (2017) Genetic population structure of a coastal marine fish (sheepshead; Archosargus probatocephalus [Sparidae]) in the southeastern United States: multiple population clusters based on species-specific microsatellite markers. Bull Mar Sci 93:691–713
Seyoum S, Puchutulegui C, McBride RS (2016) Isolation and characterization of 24 polymorphic microsatellite loci for the study of genetic population structure of sheepshead, Archosargus probatocephalus (Actinopterygii, Perciformes, Sparidae). BMC Res Notes 9:251
Stunz GW, Minello TJ, Rozas LP (2010) Relative value of oyster reef as habitat for estuarine nekton in Galveston Bay, Texas. Mar Ecol Prog Ser 406:147–159
Swofford DL (2000) PAUP*: phylogenetic analysis using parsimony (*and other methods). Sinauer, Sunderland, MA
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA7: molecular evolutionary genetics analysis v7.0. Mol Biol Evol 30:2725–2729
TPW (Texas Parks and Wildlife Department) (2005) Trends in relative abundance and size of selected finfishes and shellfishes along the Texas coast: November 1975–December 2003. Management Data Series No. 232. Texas Parks and Wildlife Department, Austin, TX
Wilcox WA (1897) Commercial fisheries of the Indian River, Florida. Report of the United States Commissioner of Fish and Fisheries 1896:249–262
Wolf DE, Takebayashi N, Rieseberg LH (2001) Predicting the risk of extinction through hybridization. Conserv Biol 15:1039–1053
Xue X, Xue J, Liu W, Adams DH, Kannan K (2017) Trophic magnification of parabens and their metabolites in a subtropical marine food web. Environ Sci Technol 51:780–789
Yu L, Luan P-T, Jin WI, Ryder OA, Chemnick LG, Davis HA, Zhang Y-P (2011) Phylogenetic utility of nuclear introns in interfamilial relationships of Caniformia (Order Carnivora). Syst Biol 60:175–187
Acknowledgements
We thank the scientists of the Fisheries-Independent Monitoring program at the Indian River, Tequesta, and St. Petersburg field laboratories of the Florida Fish and Wildlife Conservation Commission’s Fish and Wildlife Research Institute for their assistance in the collecting of samples. We are grateful to many including Mike Tringali, Brandon Barthel, Luiz Barbieri, Brent Winner, Shanae Allen, Sarah Seifu, Colin Shea, and Bland Crowder for helpful suggestions and comments on this manuscript. We are also grateful to R.G. Gilmore for providing us with information on sheepshead in the Indian River Lagoon system. This work was supported by the U.S. Department of the Interior, U.S. Fish and Wildlife Service, under the Federal Aid in Sport Fish Restoration Program, and by proceeds from State of Florida saltwater recreational fishing licenses. The statements, findings, and conclusions are those of the authors and do not necessarily reflect the views or policies of the Department of the Interior. Mention of trade names or commercial products does not constitute their endorsement by the U.S. government.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
10592_2019_1244_MOESM1_ESM.jpg
Electronic supplementary material 1—Phylogenetic relationships of four sparid species (Archosargus probatocephalus, A. rhomboidalis, Lagodon rhomboides, and Calamus nodosus) based on each of three mtDNA regions constructed with eight other closely related sparids extracted from Orrell and Carpenter (2004). Bootstrap values (1,000 replicates) are shown on the branches in % for Maximum Likelihood/MrBayes/Maximum Parsimony. Bootstrap values of only maximum likelihood are given at nodes of the extracted eight species in the mtDNA topology. Substitution models are based on the Akaike information criterion procedures in MEGA are given in Table S3. (JPG 1000 kb)
10592_2019_1244_MOESM2_ESM.jpg
Electronic supplementary material 2—Phylogenetic relationships of four sparid species (Archosargus probatocephalus, A. rhomboidalis, Lagodon rhomboides, and Calamus nodosus) based on each of three nuclear DNA introns. Bootstrap values (1,000 replicates) are shown on the branches in % for Maximum Likelihood/MrBayes/Maximum Parsimony Substitution models used for the phylogenetic analyses are given in Table S3. Crescent shapes at terminal nodes indicate collapsed branches of four or more (median eight) individuals varying between one and three base pairs. (JPG 254 kb)
10592_2019_1244_MOESM3_ESM.jpg
Electronic supplementary material 3—Distributions of assignment values (q-values) for five simulated classes of hybrids: pure sheepshead (P1), second-generation backcrosses to sheepshead (DBCP1), first-generation backcrosses to sheepshead (BCP1), first-generation hybrid between sheepshead and western Atlantic sea bream (F1), first-generation backcross to sea bream (BCP2), second-generation hybrid to sea bream (DBCP2), and pure sea bream (P2). Results are based on the simulation of 100 individual genotypes in HYBRIDLAB and the admixture algorithm implemented in STRUCTURE at K = 2. An assignment value of 1.0, or 0.95, would indicate that an individual was pure. (JPG 423 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Seyoum, S., Adams, D.H., Matheson, R.E. et al. Genetic relationships and hybridization among three western Atlantic sparid species: sheepshead (Archosargus probatocephalus), sea bream (A. rhomboidalis) and pinfish (Lagodon rhomboides). Conserv Genet 21, 161–173 (2020). https://doi.org/10.1007/s10592-019-01244-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-019-01244-7